Exploitation of local adjacencies for parallel construction of a Reeb graph variant: cerebral vascular tree case
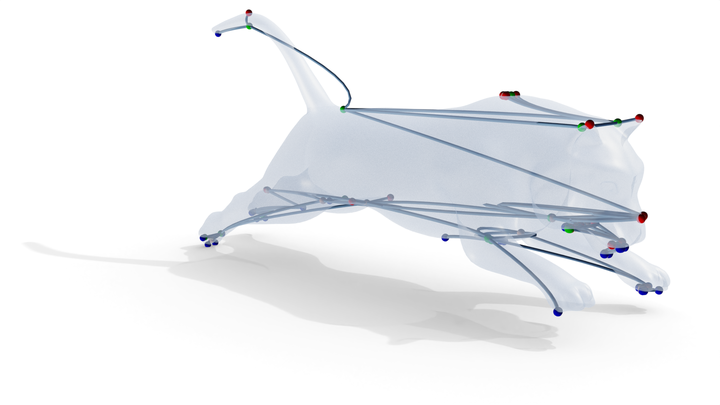 Computation of the LGRG on a tiger.
Computation of the LGRG on a tiger.
Abstract
Strokes concerned more than 795,000 individuals annually in the United States as of 2021. Detecting thrombus (blood clot) is crucial for aiding surgeons in diagnosis, a process heavily reliant on 3D models reconstructed from medical imaging. While these models are very dense with information (many vertices, edges, faces in the mesh, and noise), extracting the critical data is essential to produce an accurate analysis to support the work of practitioners. Our research, conducted in collaboration with a consortium of surgeons, leverages generalized maps (g-maps) to compute quality criteria on the cerebral vascular tree. According to medical professionals, artifacts due to noise and thin topological changes are significant parameters among these criteria. These parameters can be determined via the Reeb graph, a topological descriptor commonly used in topological data analysis (TDA). In this article, we introduce a novel classification of saddle points, and a Reeb graph variant called the Local to Global Reeb graph (LGRG). We present parallel computation methods for critical points and LGRG, relying only on local information thanks to the homogeneity of the g-map formalism. We show that LGRG preserves the most subtle topological changes while simplifying the input into a graph formalism that respects the global structure of the mesh, allowing its use in future analyses.